This paper examines the performance of different machine and deep learning algorithms in classifying colon histological images using different feature extraction methods. The relationship between the feature extraction methods and the selected machine learning methods to improve the classification accuracy is analyzed. Widely used methods like local binary patterns, histograms of oriented gradients, Gabor filter and Dobeshi wavelets are investigated for feature extraction from colon histological images. The features extracted by histogram of oriented gradients and Gabor filter methods were used as a single joint feature vector. And popular machine learning methods such as Support vector machine, Decision trees, Random forest, k-nearest neighbors and Naive Bayesian method were used to classify the selected images. The paper also investigates ensemble methods using gradient bousting and voting classifier as examples. The authors also focus on the study of convolutional neural networks as they are one of the main deep learning methods at the moment. The classification methods selected for analysis are compared in terms of classification accuracy and time taken for training and recognition. All pre-defined and adjustable parameters of both feature extraction methods and classification methods were personally selected by the authors as a result of experimental studies, which were conducted using a software tool created in the Python programming language on a set of LC25000 histological images. The software created is easily customizable and can be used in the future to investigate classification methods on other datasets.
| Published in | American Journal of Artificial Intelligence (Volume 8, Issue 2) |
| DOI | 10.11648/j.ajai.20240802.12 |
| Page(s) | 41-47 |
| Creative Commons |
This is an Open Access article, distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution and reproduction in any medium or format, provided the original work is properly cited. |
| Copyright |
Copyright © The Author(s), 2024. Published by Science Publishing Group |
LBP, HOG, Dobeshi, Feature Extraction, Classification
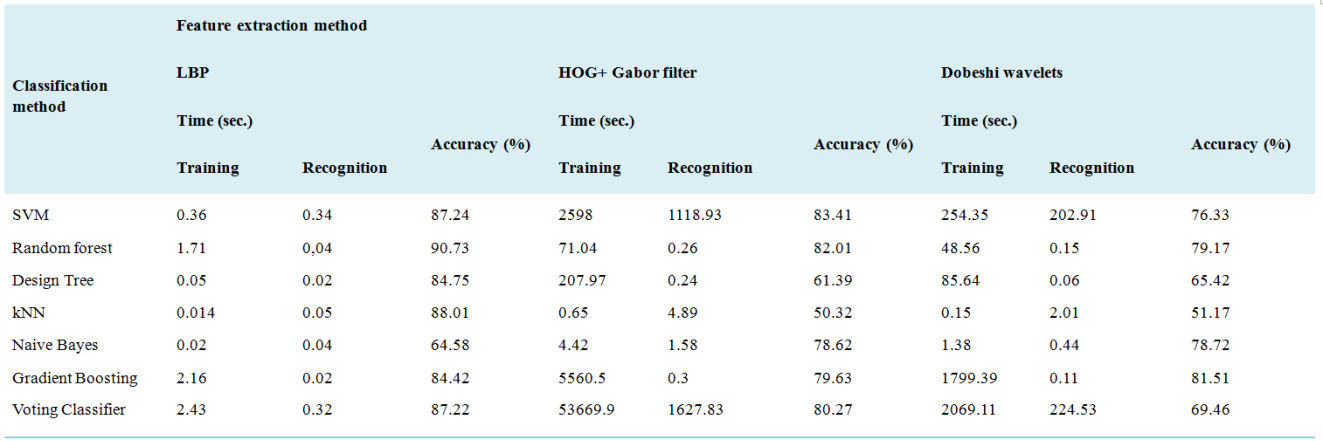
Classification method | Time (sec.) | Accuracy (%) | |
|---|---|---|---|
Training | Recognition | ||
CNN | 12936.11 | 194.7 | 98,2 |
HOG | Histograms of Oriented Gradients |
LBP | Local Binary Patterns |
CNN | Convolutional Neural Networks |
DW | Dobeshi Wavelets |
DWT | Discrete Wavelet Transforms |
SVM | Support Vector Machine |
kNN | k-Nearest Neighbor |
| [1] | Gurcan, M. N., Boucheron, L. E., Can, A., Madabhushi, A., Rajpoot, N. M. and Yener, B. Histopathological image analysis: A review. In IEEE Reviews in Biomedical Engineering, 2009, vol. 2, pp. 147-171. |
| [2] | De Matos, J., Ataky, S. T. M., de Souza Britto Jr, A., Soares de Oliveira, L. E. and Lameiras Koerich, A. Machine learning methods for histopathological image analysis: A review, Electronics. 2021, 10(5), 562. |
| [3] | Aeffner, F., Zarella, M. D., Buchbinder, N., Bui, M. M., Goodman, M. R., Hartman, D. J., Lujan, G. M., Molani, M. A., Parwani, A. V., Lillard, K. and Turner, O. C. Introduction to digital image analysis in whole-slide imaging: a white paper from the digital pathology association, Journal of pathology informatics. 2019, 10(1), p. 9. |
| [4] | Mutlag W. K. et al. Feature extraction methods: a review, Journal of Physics: Conference Series. – IOP Publishing, 2020. – Т. 1591. – №. 1. – С. 012028. |
| [5] | Dalal, N., Triggs, B. Histograms of oriented gradients for human detection. In IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05), 2005, San Diego, CA, USA, pp. 886-893 vol. 1. |
| [6] | Manjunath, B. S. and Ma, W. Y. Texture features for browsing and retrieval of image data. In IEEE Transactions on pattern analysis and machine intelligence 1996. 18(8), pp. 837-842. |
| [7] | Mallat SG. A theory for multiresolution signal decomposition: the wavelet representation. In IEEE transactions on pattern analysis and machine intelligence 1989. 11(7), pp 674-693. |
| [8] | Ojala, T., Pietikäinen, M., Harwood, D. A comparative study of texture measures with classification based on featured distributions. Pattern recognition. 1996, 29(1), pp. 51-59. |
| [9] | LeCun, Y., Bengio, Y., Hinton, G., 2015. Deep learning. Nature, 521(7553), 436-444. |
| [10] | Wyld, Henry William, and Gary Powell. Mathematical methods for physics. CRC Press, 2020. |
| [11] | Borkowski, A. A., Bui, M. M., Thomas, L. B., Wilson, C. P., DeLand, L. A. and Mastorides, S. M., 2019. Lung and colon cancer histopathological image dataset (lc25000). arXiv preprint arXiv:1912.12142. |
APA Style
Mirzaev, N., Meliev, F. (2024). Investigation of Histological Image Classification Methods Using Different Feature Extraction Techniques. American Journal of Artificial Intelligence, 8(2), 41-47. https://doi.org/10.11648/j.ajai.20240802.12
ACS Style
Mirzaev, N.; Meliev, F. Investigation of Histological Image Classification Methods Using Different Feature Extraction Techniques. Am. J. Artif. Intell. 2024, 8(2), 41-47. doi: 10.11648/j.ajai.20240802.12
AMA Style
Mirzaev N, Meliev F. Investigation of Histological Image Classification Methods Using Different Feature Extraction Techniques. Am J Artif Intell. 2024;8(2):41-47. doi: 10.11648/j.ajai.20240802.12
@article{10.11648/j.ajai.20240802.12,
author = {Nomaz Mirzaev and Farkhod Meliev},
title = {Investigation of Histological Image Classification Methods Using Different Feature Extraction Techniques
},
journal = {American Journal of Artificial Intelligence},
volume = {8},
number = {2},
pages = {41-47},
doi = {10.11648/j.ajai.20240802.12},
url = {https://doi.org/10.11648/j.ajai.20240802.12},
eprint = {https://article.sciencepublishinggroup.com/pdf/10.11648.j.ajai.20240802.12},
abstract = {This paper examines the performance of different machine and deep learning algorithms in classifying colon histological images using different feature extraction methods. The relationship between the feature extraction methods and the selected machine learning methods to improve the classification accuracy is analyzed. Widely used methods like local binary patterns, histograms of oriented gradients, Gabor filter and Dobeshi wavelets are investigated for feature extraction from colon histological images. The features extracted by histogram of oriented gradients and Gabor filter methods were used as a single joint feature vector. And popular machine learning methods such as Support vector machine, Decision trees, Random forest, k-nearest neighbors and Naive Bayesian method were used to classify the selected images. The paper also investigates ensemble methods using gradient bousting and voting classifier as examples. The authors also focus on the study of convolutional neural networks as they are one of the main deep learning methods at the moment. The classification methods selected for analysis are compared in terms of classification accuracy and time taken for training and recognition. All pre-defined and adjustable parameters of both feature extraction methods and classification methods were personally selected by the authors as a result of experimental studies, which were conducted using a software tool created in the Python programming language on a set of LC25000 histological images. The software created is easily customizable and can be used in the future to investigate classification methods on other datasets.
},
year = {2024}
}
TY - JOUR T1 - Investigation of Histological Image Classification Methods Using Different Feature Extraction Techniques AU - Nomaz Mirzaev AU - Farkhod Meliev Y1 - 2024/08/20 PY - 2024 N1 - https://doi.org/10.11648/j.ajai.20240802.12 DO - 10.11648/j.ajai.20240802.12 T2 - American Journal of Artificial Intelligence JF - American Journal of Artificial Intelligence JO - American Journal of Artificial Intelligence SP - 41 EP - 47 PB - Science Publishing Group SN - 2639-9733 UR - https://doi.org/10.11648/j.ajai.20240802.12 AB - This paper examines the performance of different machine and deep learning algorithms in classifying colon histological images using different feature extraction methods. The relationship between the feature extraction methods and the selected machine learning methods to improve the classification accuracy is analyzed. Widely used methods like local binary patterns, histograms of oriented gradients, Gabor filter and Dobeshi wavelets are investigated for feature extraction from colon histological images. The features extracted by histogram of oriented gradients and Gabor filter methods were used as a single joint feature vector. And popular machine learning methods such as Support vector machine, Decision trees, Random forest, k-nearest neighbors and Naive Bayesian method were used to classify the selected images. The paper also investigates ensemble methods using gradient bousting and voting classifier as examples. The authors also focus on the study of convolutional neural networks as they are one of the main deep learning methods at the moment. The classification methods selected for analysis are compared in terms of classification accuracy and time taken for training and recognition. All pre-defined and adjustable parameters of both feature extraction methods and classification methods were personally selected by the authors as a result of experimental studies, which were conducted using a software tool created in the Python programming language on a set of LC25000 histological images. The software created is easily customizable and can be used in the future to investigate classification methods on other datasets. VL - 8 IS - 2 ER -